HSC Biology Syllabus Notes
Module 5 / Inquiry Question 4
Overview of Inquiry Question #1
Learning Objective #1 – Predicting variations in offspring’s genotype by modelling meiosis – crossing over, fertilisation and mutation
Learning Objective #2 – Model the formation of new combinations of alleles during meiosis and sexual reproduction – autosomal, sex linkage, co-dominance, incomplete dominance and multiple alleles inheritance
Learning Objective #3 – Constructing and interpreting Punnett Squares
Learning Objective #4 – Constructing and interpreting Pedigrees
Learning Objective #5 – Examining and monitoring the frequency of characteristics in a population in order to identify trends, patterns, relationships and limitations in data
Allele frequency, genotype frequency
Trait frequency such as phenotype
Learning Objective #6 – Exploring single nucleotide polymorphism
NEW HSC Biology Syllabus Video – Genetic Variation
Week 4 Homework Questions
Week 4 Curveball Questions
Week 4 Extension Questions
Solutions to Week 4 Questions
Learning Objective #1 - Predicting variations in offspring's genotype by modelling meiosis - crossing over, fertilisation and mutation
Genetic Variation created by crossing over
Crossing over is process involving the exchange of corresponding gene segments of non-sister chromatids between homologous chromosome pairs (double-stranded chromosome pairs for the most organisms).
This effectively creates creates new allele combinations, known as recombination.
It is important to stress that stating ‘new allele combinations being created as a result of crossing over’ is critical in HSC Biology as it is what appears on the marking criteria.
Below is a diagram illustrating the crossing over process between one pair of homologous chromosomes.
Notice that the crossing over occurs between non-sister chromatids, one from each double-stranded chromosome of the homologous pair. Each parent of the organism contribute one double-stranded chromosome.
For example, perhaps the blue one can be from the father (paternal chromosome) and the pink one can be from the mother (maternal chromosome).
Chiasma is the point where crossing over occur. Plural is chiasmata. There can be multiple chiasmata, usually the longer chromosome, the more points of crossing over. The chiasma the visible component of the homologous chromosomes during crossing over in Prophase I in meiosis I.
Recall from Week 2’s notes that crossing over does not occur in mitosis because the homologous chromosomes are NOT aligned side-by-side along the equator of the cell to allow overlapping segments of non-sister chromatids along the equator in the nuclear membrane.
Fun Fact: Crossing over can in fact occur between sister chromatids in one double-stranded chromosome (with requiring another double-stranded chromosome). However, it is ONLY crossing over between non-sister chromatids in a homologous pair where where new allele combinations are created.
You may be wondering why?
Well, if you recall that sister chromatids are exact copies of each other, this means that the alleles on sister chromatids (coding for the any of the genes) are identical! Take a look at the diagram above. Each red dashed line highlights a locus on each chromatid.
Prior to crossing over, each locus (position) on each sister chromatid has the same allele.
For example, take the blue double-stranded chromosome, both of the sister chromatids have the ‘B’ allele that codes for black hair colour. Now, look at the other double-stranded chromosome (pink) in the homologous pair. At the locus of both sister chromatids that carries the gene that codes for eye colour, they both have the ‘b’ allele that codes for blue eye colour.
NOTE: In this example, ‘B’ is an allele means that it codes for black eye colour and ‘b’ is an allele codes for blue eye colour. The letters (or alleles to be exact) can code for different colours or genes depending on the question given to you on the day. For instance, in the exam, the question will tell you that what gene each letter codes for AND what the capital and lowercase of each letter would mean in terms of alleles.
By convention, capital letter represents a dominant allele and lowercase represents a recessive allele. We will talk about dominant and recessive alleles soon in this week’s notes.
Just to enhance your prior knowledge from Week 2: As you can see from the diagram shown above, the blue and red double-stranded chromosomes are called homologous pairs because, at the each locus (each locus shown by a red dashed line) of the chromosomes in the homologous pairs there are alleles that code for the same gene.
Fun Fact: The longer the distance genes are separated along each of the chromosome (chromatid to be precise), the greater the chance of crossing over. Generally, this will depend on the length of the chromosome. The greater the length, the more chance of crossing over and more chiasmata will exist during crossing over.
In the previous diagram, I have drew light green arrows on the homologous chromosomes after crossing over to indicate the alleles that belong to each of the four chromatids.
If I were to separate the four chromatids in the previous diagram on its own, the resulting allele combinations for the four chromatids would be as follows:
As you can see, there are two NEW allele combinations that has been created as a result of crossing over.
These new allele combinations are bHC and BhC which did not exist prior to crossing over.
Extra crossing over scenario that is BEYOND the HSC Biology Course
(You WILL NOT be asked about this in HSC Biology Exams)
Note that this diagram illustrating crossing over process, before and after, is slightly different to the previous diagrams. Can you spot the difference?
The difference is that the chiasma, i.e. the point of crossing over, is NOT located between the genes that codes for eye colour and height unlike in the previous diagram. As you can see in the products after crossing over, there is still new allele combinations formed. However, there is one consequence.
This type of crossing over usually is hard to detect (often undetected by normal means) compared to the previous crossing over scenarios. This means that the average number of crossing over detected would be lower than the true amount of crossing over that actually happened.
Scientists often perform crossing over experiments in attempt to determine the locus of chromosome (and thus DNA) that holds alleles that codes for a particular gene.
So, having undetected crossing over event that in fact actually occurred would yield inaccurate results.
This in fact relates to single nucleotide polymorphism which we will be covering as the last learning objective in this week’s notes.
Significance of crossing over
The process of crossing over creates new allele combinations adds diversity of the gene pool of the population as the four gametes formed in meiosis will not be identical but rather can have different an allele for each gene. Also, upon fertilisation, two random gametes with some variation in their alleles for certain genes will combine and produce an offspring with an unique allele combination to their parents. This therefore increases genetic variation in a population and can introduce new phenotypes that potentially* could be expressed.
Genetic variation in a population provides a pathway for evolution to occur. Without genetic variation, there will be no mechanisms for evolution. Genetic variation can be introduced into a population’s gene pool via:
Sexual reproduction (crossing over, independent assortment, random segregation and fertilisation) or
Mutation
*The word ‘potentially’ is used because whether or not the allele will be expressed will depend on whether or not it is recessive and dominant which also depends on the complementary allele for the same gene.
Dominant and Recessive Alleles
Alleles are alternative (or different) versions of a gene that differ by their DNA sequence but codes for a protein that responsible for a same trait (e.g. hairy or hairless). An allele can be classified as dominant or recessive.
Dominant alleles are always expressed over recessive alleles if they are both present in the genotype for a gene of the organism.
By convention, dominant alleles are denoted with capital letters.
Recessive alleles are denoted with lower-case letters.
If the the alleles are coding for the same trait, the same letter is used. If not, different letters are used.
For example, in the diagrams shown previously, for the gene that codes for hair colour, there are two types of alleles, B and b.
The ‘B’ allele would be the dominant allele for hair colour and b would be the recessive allele for hair colour. In HSC Biology, the question will tell you what the hair colour for dominant allele as well as for the recessive allele for hair colour.
Some question requires you to determine what trait each allele expresses and whether or not they are dominant or recessive, in that case, the question will give you sufficient information to allow you to work it out. In that case, you will need to use Punnett Squares which we will learn this week. There will be these questions in this week’s homework set to allow you to practice. Give it a go. Solutions will be uploaded soon.
Anyhow, below are some rules of thumb about dominant and recessive allele combinations:
If the organism (parent and/or offspring) has two dominant allele for a particular gene, the dominant allele will be expressed.
If the organism (parent and/or offspring) has two alleles for a particular gene, one dominant and the other is recessive, the dominant allele will be expressed.
If the organism (parent and/or offspring) has two recessive alleles for a particular gene, the recessive allele will be expressed.
Fertilisation
As illustrated in previous diagram, ‘Four different combinations of alleles after crossing over’, each chromatid begin segregating in Anaphase II and be completely segregated into a different gamete after Telophase II.
This occurs for all 23 pairs of homologous chromosome (one pair being sex chromosomes, XX or XY).
In the previous diagrams , we only drew two chromosomes (a homologous pair) that are crossing over just to satisfy the purpose of a simplistic illustration of the crossing over of chromosomes in a homologous pair.
As you may know now, when the gametes fuse together (egg and sperm except for fauna), a zygote is formed which has a diploid number of chromosomes. That is, the zygote has 46 chromosomes (23 homologous pairs).
So, in meiosis, the process of crossing over effectively creates the genetic variation in the gametes due to their difference in alleles for various genes (as we have mentioned earlier). Also, the random process of fertilisation between gametes (egg and sperm) also give rise to the genetic variation in the zygote whereby the two gamete have different alleles such that the zygote will have different allele combinations than its parents.
Due to meiosis, each gamete will have one allele that codes for a particular gene. Upon fertilisation where the female and male gametes fuse together to form a zygote, the zygote will then two alleles that code for each gene (e.g. gene for eye colour).
Therefore, the process of fertilisation creates new genotypes for the offspring that are, for the most part, different from each individual parent.
Note that if both parents the homozygous dominant for the same gene. Homozygous dominant just means that the individual has two same alleles (homozygous) and the alleles are dominant (capital letter). So, if both parents have homozygous dominant alleles for a trait, then the offspring will have genotype that is exactly the same as the parent (homozygous dominant too) though ONLY for that particular gene. It is unlikely that both parents will have EXACTLY the same genotype (allele combinations) for EVERY GENE. In that case, they be twins. LOL.
We will further explore and talk about homozygous dominant later when we look at Punnett Squares later in this week’s notes.
Also note, in reality, most traits (e.g. height) are governed by more than one gene.
Recall that independent assortment and random segregation both facilitate in increasing the genetic variation in offsprings of the new generations. Although they do not create new allele combinations, they do introduce variations in the way which alleles that specifies different genes on non-homologous chromosomes are assorted independently relative to each other (independent assortment) and eventually different chromatids that carries different alleles for certain genes can be segregated into different gametes (random segregation).
This means that these two independent assortment and random segregation introduce (genetic) variation into the alleles that each gamete inherit after Cytokinesis II in Telophase II.
The specifics in how independent assortment and random segregation increases the genetic variation in the offspring was discussed in Week 2’s note. If required, please revisit that section to refresh memory.
Mutation
Recall that crossing over introduces new combinations of alleles that already exist in the chromatids by exchanging corresponding gene segments between non-sister chromatids.
Mutation, however, alters the allele’s identity because the DNA sequence is altered. This new DNA sequence can lead to an alternative expression of the gene (e.g. blue eye colour instead of black eye colour), hence new allele (different DNA sequence for same gene) is created.
In most cases, mutation only involves the modification of a DNA nucleotide, changing its nitrogenous base (point mutation).
In some more extreme cases, it can modify the DNA sequence of a portion of chromosome that involves one or more genes (chromosomal mutation).
We will explore point mutation and chromosomal mutation in Module 6 – Inquiry Question 1.
Anyhow, after learning about protein-synthesis in last week’s notes, where the creation of mRNA uses the DNA sequence of a gene as a template, it is clear that altering the nucleotide sequence of a gene will result in the specification of a different protein OR may modify the structure and function of the protein such that it will no longer functions as efficiently or completely not functional at all.
Suppose, you have an allele that codes for blue eye colour. This allele can be exchanged with another non-sister chromatid during crossing over.
However, mutation completely alters the identity of this allele originally coded for blue eye colour. This means that it may code for something slightly or completely different instead. Perhaps maybe, green eye colour.
Since alleles are altered, this would mean that through mechanisms of sexual reproduction and fertilisation, mutated allele will be inherited by gamete and possible give rise to offspring when fertilised. This would mean that a new allele will be introduced into the population. This would mean that there would be an increase in genetic variation in the population as allele frequency will increase.
Note that mutation on genes in autosomes and sex chromosomes will be passed onto offspring if ONLY the mutation occurs in the parents’ germ cell. The gametes derived from the parent germ cell then becomes an offspring when gamete is fertilised where the offspring can inherits the mutation. Most mutations passed onto offspring may not expressed as they could be recessive so offspring needs to inherit one mutated allele from each parent in order to express the mutated characteristic.
Whether or not the zygote will express the new allele due to mutation will depend on the zygote’s genotype (dominant or recessive for that gene) which we will explore very shortly when we deal with Punnett Squares.
If the mutated allele is expressed, it is important to consider whether or not the expressed trait that it codes for will become an improvement or hinderance to the organism’s daily activities.
Mutations are permanent changes to an organism’s DNA sequence.
Mutation and its implications will be discussed in greater detail in Module 6.
Mutation is considered the principal and original source in creating genetic variation in a population. This is because they provide the creation of new alleles by altering DNA sequence. The parent can be copy such mutated DNA via meiosis I which gametes and, when fertilised, the offspring can inherit such mutated DNA.
Not all of the four gametes must inherit such mutated DNA though.
Anyways, similar to crossing over, independent assortment, random segregation, we should now know that mutation also creates genetic variation in a population which provides a pathway for evolution to occur.
Without genetic variation, there will be no mechanisms for evolution. This ties nicely to support your responses when asking questions regarding ‘ensuring’ the continuity of species as per inquiry question one.
As genetic variability in a population increases, the species’ population ability to adapt to its environment over time also increases. It is through such adaptability of species in a population overtime whereby we witness evolution due to shifts in dominant or new characteristics in the species’ population.
Learning Objective #2 - Model the formation of new combinations of genotype produced during meiosis, including and interpreting examples of:
- Autosomal Inheritance
- Sex-Linkage Inheritance
- Co-dominance Inheritance
- Incomplete Dominance Inheritance
- Multiple alleles Inheritance
An autosome is a chromosome in an organism that is not a sex chromosome.
In humans, we have 22 pairs of autosomes and one pair of sex chromosomes. Genes are present in both autosomes and sex chromosomes.
The process of transferring genes (DNA) present in the parents’ autosomes to offspring is called autosomal inheritance.
The process of transferring genes (DNA) present in the parents’ sex chromosomes to offspring is called sex-linked inheritance or X-linked inheritance.
The genes that is present in autosomes and sex chromosomes, when passed to offsprings, exhibit different inheritance patterns or combinations of genotypes in terms of phenotype.
More specifically, the likelihood of an offspring exhibiting a recessive trait that is passed on autosomal inheritance is equal for both males and females. However, the likelihood of offspring exhibiting a recessive trait that is passed on via sex chromosomes inheritance is greater for males than in females.
Let’s have a look at why this is the case by exploring both modes of inheritance.
Autosomal Inheritance
Recall that in humans, there are two alleles for a given gene where the alleles help determine the trait of organism (both the parents and offspring).
For illustration purposes, let’s use the gene that codes for eye colour. The gene that codes for eye colour will be present in both the female and male.
Just to get a clearer picture and as a recap, do recall that a gene is basically a segment of a DNA or a sequence of DNA nucleotides. Chromosomes are made up of DNA (wrapped around proteins called histones) for eukaryotes. Approximately 40% of a chromosome is made up of DNA and the other 60% is made up of proteins.
Now, let’s return to our example. So, each parent will have two alleles for its eye colour gene. Suppose that:
The mother has an allele that codes for blue eye colour and an allele that codes for black eye colour.
The father have both alleles that code for black eye colour.
Let’s also suppose that the allele that codes for black eye colour is denoted by ‘B’ and the blue eye colour is denoted by ‘b’.
As mentioned earlier, the dominant gene is denoted by a capital case of the letter and the recessive gene is denoted by the lower case version of the same letter. Therefore, in our case, black eye colour is a dominant gene whereas blue eye colour is a recessive gene.*
* In the exam you could either be told which is allele is dominant or recessive through the use of capital and lower case letters. If not, you will be told by words e.g. “Black eye colour is a dominant gene whereas blue eye colour is a recessive gene.” In that case, since no letter is given to you, you will need to chose a (same) letter to denote both alleles where the lower case letter represents the recessive allele and capital letter for the dominant allele.
Make sure you write down in words that letter you chose to use represent for that particular gene as well as what lower case and capital letter means in terms of recessive/dominant alleles and what characteristics they represent (e.g. black/blue eye colour).
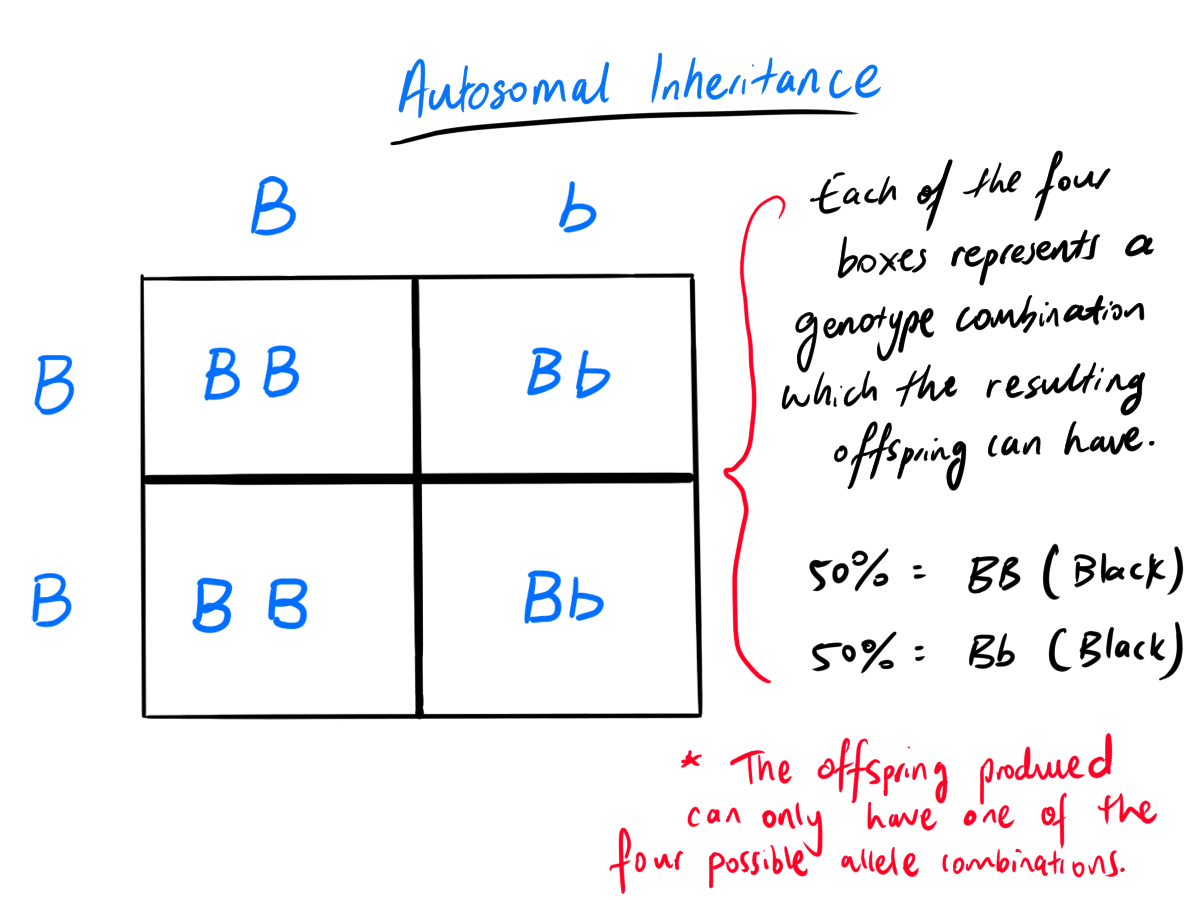
Let’s have a look at how the alleles coding for eye colour can be passed on via the crossing over of the mother and father gametes. To do this, one method is to construct a Punnett Square.
Therefore, in our example, the mother will have the genotype, Bb, which is crossed over with the father with genotype, BB.
It does not matter which side (top or left of the square) you write the genotype for the male or female.
The order of the alleles also does not matter, e.g. (Bb or bB for the mother).
Each letter represents the allele for that particular gene (in this case gene for eye colour) that the parent inherited. Since each gamete only inherits one allele for each gene from each parent, you could think that each letter is essentially a gamete if you wish.
Recall that each parent has two alleles for the eye colour and it can pass on one of the two alleles to its offspring (B or b).
In the above example, the mother has B and b alleles for eye colour. So, she can pass on either the B or b allele to each of its gametes (egg cells) during meiosis. This is shown on the left of the Punnett Square.
For the father, he only two B alleles. So, he can only pass on B alleles to its gametes (sperm cells). This is shown on top of Punnett Square.
Don’t forget that each parent can produce many, many of gametes during his or her or its lifetime.
So, the Punnett Square therefore considers all the possible alleles which the parent have and could potentially pass onto its gamete and crosses these alleles together to depict the different allele combination for a particular gene which the offspring can inherit from both parents. The Punnett Square allows us to get four genotype combinations (allele combinations for a gene) as shown in the four boxes in the Punnett Square above. Some of the genotype combinations can be identical to each other as shown in the diagram.
When the mother and father crosses over, the resulting offspring will inherit one allele from each parent. There are four different possible genotype combination for the offspring’s eye colour. These are BB, Bb, BB and Bb. This means there are 50% chance that the offspring will have the genotype – BB. Also, there is 50% chance that the offspring will have genotype Bb.
Recall that the B allele is dominant over b allele (hence the capital letter). This means that although the offspring can inherit one of the two different genotype combinations, the resulting offspring only has one phenotype combination! That is, the offspring will have a 100% probability to have a black eye colour regardless of which genotype combination (BB or Bb) that its inherits from the parents.
In the exam, it is important for you to write in your response – either below or next to your Punett Square diagram, the following:
Genotype:
50% = BB
50% = Bb
Phenotype:
100% = Black eye colour
It is important to know that when both alleles for a gene of a parent are identical, the parent is called homozygous for that particular trait or gene. If the parent’s alleles for a gene are different, the parent are called heterozygous for that gene or trait. The same concept applies to offsprings.
Please DO NOT get the terms ‘homozygous’ and ‘homologous’ confused!
Homologous was used to refer to chromosomes pairs that have the carries genes at every corresponding locus. This is different to homozygous which refers to having different alleles of a particular trait.
If the organism contains one dominant allele and one recessive allele for a certain gene, it is called heterozygous dominant. For example, Bb.
If the organism contains two dominant alleles for a certain gene, it is called homozygous dominant. For example, BB.
Since there is only one case of recessive alleles, that is bb, it is just called recessive. Recessive is always homozygous.
Sex-Linkage or X-Linked Inheritance
Recall that earlier in this week’s note, we say that males are more likely to exhibit traits that are sex-linked inherited compared to females.
Suppose that you know of a disease where the disease exhibited more often in males and female such as colour-blindness, this would suggest that the alleles responsible for colour-blindness would be carried by sex chromosomes rather than autosomes as outlined earlier. We will have a look why by examining how a sex-linked disease such as colour-blindness can be passed on from parents to the offspring through allele inheritance.
Sex-linked traits are characteristics whereby the alleles responsible for them are only located on the X chromosome. This means that there is no allele responsible for the sex-linked trait on the Y chromosome. Only male gametes have Y sex chromosomes.
The alleles that are responsible for expression sex-linked traits are recessive.
This means that males only require one of the recessive allele for the sex-linked trait to be expressed as there is no second allele on the Y chromosome to allow the overriding of the recessive allele on the X chromosome in males.
Females, however, have two X sex chromosomes. In each chromosome, there is one allele that codes for the sex-linked trait (for example having colourblindness or normal colour vision). If one of the female’s X chromosome contains the recessive allele responsible for the colourblindness and the other X chromosome contains the dominant allele (normal colour vision), the female will not be colour-blind.
This is because the dominant allele from one of the X chromosome overridden the recessive allele that is responsible for expressing colour-blindness located on the other X chromosome.
As males only have an allele on its X chromosome and not on its Y chromosome, males are more likely to express sex-linked traits than females. This is because males only need one allele that codes for colour-blindness and the male will have the recessive sex-linked trait. For females, both recessive alleles must be inherited in order for a female to have the recessive sex-linked trait.
Notice in the above Punett Square diagram that we included genders of the parent, represented by their sex chromosomes, to show where alleles are carried on the sex chromosomes. Showing the sex chromosomes or gender is crucial to indicate to the marker that we are dealing with sex-linked inheritance.
This is important because the offspring’s gender (more specifically offsprings’ sex chromosomes) plays a role in determining whether or not the offspring will express the sex-linked trait such as colour-blindness.
Notice that in all scenarios when a male and female gamete (coming from father and mother respectively) is combined to form a zygote, there is equal chance of the offspring (zygote) being a male and female. This is consistent to reality.
Also notice that all males have a XY sex chromosome pair. All females have a XX sex chromosomes pair.
These sex chromosomes are within the male and female gametes of the parents.
Let’s explore at of the above scenarios, one at a time. Suppose that the allele is responsible for colour-blindness is denoted by a. The allele that is responsible for normal colour vision is denoted by A.
In scenario 1, as shown in the diagram, both the father’s and mother’s sex chromosomes do not have (i.e. do not carry) the allele responsible for colour-blindness. When the father and mother gametes crosses over, the offspring can either be female (XX) or male (XY) with equal chance. As the recessive colour-blindness allele is not present in either of the parents’ sex chromosomes, the offspring will not have genotype containing the colour-blindness allele. Therefore, the offspring that is born will not have colour-blindness regardless of it is male or female.
In scenario 2, as shown in the diagram, the father is affected by colour-blindness personally. That is, he is colour-blind. The mother is not colour-blind, nor she is a carrier of the allele responsible for colour-blindness. When the father and mother gametes combines to form the zygote, the offspring that develops can be either male or female with equal probability (as per usual). If the offspring is a female, then it will be a carrier of the recessive allele responsible for colour-blindness. That being said, all female offsprings will not be colour-blind themselves. Any female offspring produced will only be carriers of the allele responsible for colour-blindness.
In scenario 3, as shown in the diagram, the mother is a carrier of the recessive colour-blind allele whereas the father have an allele that codes for normal colour vision. When the two crosses over, if resulting offspring is male, the male offsprings will have a 50% chance to be colour-blind. All females offsprings will only be carriers of the recessive allele responsible for colour-blindness.
Scenario 4 (not included in diagram): If the father is colour-blind and the mother is a carrier, then 50% of all male offsprings will be colour-blind. For female offsprings, they have a 50% chance of suffering from colour-blindness. The other 50% for females will be carriers of the recessive allele responsible for colour-blindness.
Scenario 5 (not included in diagram): If the mother is affected then all male offsprings will be affected regardless of the father being colourblind or not.
So what is implication of Scenario 2?
Well, since the Y sex chromosomes (which is only present in males only) do not carry any alleles, if the female offsprings in scenario 2 crosses over with a male without colour-blindness, the resulting male offsprings in the new generation will have a 50% probability of suffering from colour-blindness! On the other hand, if the resulting offspring is female, they will have a 0% probability of suffering from colour-blindness.
What will happen if the father and mother both carry the colour-blindness allele and father and mother crosses over?
Well, 50% of the female offsprings will be colour-blind as the recessive allele is expressed. This is because 50% of the females will be carriers. There is also another 50% of female offsprings where both of the X sex chromosomes for the female will carry the recessive allele responsible for colour-blindness. If the female offspring inherits this combination of allele, this would mean that the female will be colour-blind.
If the offspring is a male, he will be colour-blind with 100% chance.
Conclusion: For traits that passed on via sex-linked inheritance, males have higher chance of exhibiting the sex-linked trait compared to females as the Y chromosome does not carry the relevant allele for the trait.
NOTE: It is important to remember that, at the end of the day, the allele combinations shown on the Punnett Square are all percentages used to indicate the probability of the offspring produced of getting a particular genotype (or allele combination). An offspring can inherit a genotype that has 25% probability and not a genotype that has 75% probability.
Co-dominance Inheritance
Co-dominance occurs when both alleles for a trait are expressed at the same time without blending of the trait that each allele specifies. This occurs when there is no recessive alleles for the trait.
An example of co-dominance is when a cow have both red and white colour patches on its skin. These cows are known as roan cows. Roan cows comes from the crossing over (mating) of red and white parent cows.
The offspring that have exhibit co-dominance must be heterozygous! They have different alleles for the same gene.
Let’s suppose you have a homozygous red female cow (RR) and a homozygous white male cow (WW). If these two cows are crossed, the cow offspring will 100% be roan (RW) as shown in the diagram below.
That is, the roan cow offspring will express both alleles where one codes for red coat colour and the other with white coat colour. This is because both alleles that codes for red and white colour patches are dominant, hence the term, co-dominance.
NOTE: Not all traits are co-dominant traits! Only in co-dominant traits where both alleles will be expressed as there is no dominant and recessive relationship between the alleles that govern the co-dominant trait.
Do note that if both the crossed-over parents are both heterozygous (both roan cows), it is possible to have a roan cow offspring. However, the probability is not 100%. It will only be 50%.
This will be made more clear if you draw a Punnett square showing the crossing of two parent roan cows that are heterozygous, i.e. Rw x Rw.
Incomplete Dominance Inheritance
Incomplete dominance is similar to co-dominance in that both alleles for a given gene are expressed meaning that neither of the two alleles have complete dominance over the other. However, for traits that are incomplete dominant, there is a blending of the two parents’ phenotypes which that visible in the offspring’s phenotype.
NOTE: ‘Blending’ could refer to the blending of the genetic material (alleles). However, alleles responsible for incomplete dominance inheritance do not blend. Only the phenotypes blend during incomplete dominance. This is because two recessive alleles have the chance to be inherited and exhibited (expressed) in the subsequent generations.
In the case co-dominance, there is no blending!
An example of incomplete dominance can be found in snapdragon flowers. Suppose you have two snapdragon plants. One is a homozygous white snapdragon (WW) and the other is a homozygous red snapdragon (RR).
If you do the Punnett square crossing, you will find that resulting snapdragon offspring will have a 100% chance to have the genotype combination, RW. This genotype (RW) will give the phenotype of pink-coloured snapdragons. The red allele is not capable of coding sufficient red pigment for the snapdragon flowers to have a complete red colour. Vice versa, the white allele is not capable for coding sufficient white pigment for the snapdragon flower to have a complete white colour.
As pink colour is a blend between red and white, this is a case of incomplete dominance. Similar to co-dominance, the offspring that exhibits phenotype blending (incomplete dominance) is heterozygous.
Sometimes, you may see one form of the allele for the gene be written as W’ W’ instead of WW where W’ W’ means homozygous red and WW means homozygous white. The apostrophe is used to differentiate between the two alleles, i.e. W allele and the W’ allele when the same letter for both alleles are used.
It is up to you to use different letters or same letters (with apostrophe) in the case of co-dominance and incomplete dominance only. Just be clear on the day of exam by communicating to the marker what your symbols mean. I personally use WW to indicate homozygous white and RR to indicate homozygous red as such expressions of alleles involved incomplete dominance are by far more common from personal experience.
Another example of incomplete dominance is the different pitch levels of the human voice – low, intermediate and high pitch voices.
Multiple Alleles Inheritance
For some traits, there may be multiple alleles that code for such traits. So far, we have discussed traits that are expressed based on two alleles as examined by the scientist, Gregor Mendel.
For HSC purposes, an organism’s trait can only be governed by two alleles as only two alleles can be present at a time within an organism’s genotype. Therefore in multiple alleles inheritance, the entire population of species is examined.
For example, there are three alleles that governs the human blood type. These are O, A and B.
These alleles creates the blood types – A, B, AB and O.
An organism can inherit any two of the three alleles (A, B or O) as long as the alleles are available in the mother’s and female’s genotype.
Depending on which of the two blood type alleles an organism carries, the genotype will determine the organism’s blood type. In other to consider the third allele, the whole species population must be examined and not just the organism itself. By considering the alleles in the total population, we remove bias in examining the frequency of the allele and its presence.
The alleles coding for A and B blood types are co-dominant. This would mean that if the offspring inherits an A allele from one parent the B from another, the offspring will have the blood type AB rather than blood type A or B (which occur if the alleles have a dominant-recessive relationship).
The allele that codes for blood type O is recessive. This means that the offspring must have two alleles that codes for blood type O in order to have blood type of O. This is because if the offspring has an allele that codes for blood type A or B, the allele will override the recessive allele that codes for blood type O.
Additional information that is NOT necessary for HSC Biology: The letter to denote the gene for blood type (or more specifically, the locus for the blood type gene on the chromosome) is typically expressed as I (capital i). The alleles which are coding for different blood types are written as superscripts.
Pedigrees
A pedigree is a chart, similar to a tree diagram, that illustrates all the different phenotypes (and sometimes genotypes) of individuals in a family across several generations.
For HSC Biology exam questions, you could be given a pedigree diagram for you to determine the type of inheritance for a certain gene that codes for trait. You may also be asked to draw a pedigree diagram.
This means that you will need to know how to interpret pedigrees. There is no secret formula to instantly know what type of inheritance for a given trait.
However, there is a process which you can follow! First, select a type of inheritance that you believe is the method whereby the trait of concern is inherited. Next, look for patterns on the pedigree to verify whether or not the type of inheritance you selected or guessed is true!
If not, select another type of inheritance and look for patterns that matches that type of inheritance. Repeat until you find a pattern that matches the type of inheritance that you selected.
So now, all you need to know is the type of patterns that you could see for different types of inheritance! Let’s explore them!
There are generally four types of methods of inheritance that you need to spot patterns for when doing pedigree question.
Don’t be overwhelmed by the following patterns for each inheritance. There will be questions at the end of this week’s notes. You can give them a go and look at the solutions after where I go through step-by-step how to determine the mode of inheritance using patterns in a pedigree!
Autosomal Dominant Inheritance:
The trait have equal chance to appear on both female and male.
The trait CANNOT skip a row (each row = a different generation) on the pedigree tree diagram.
Offsprings’ that expresses the dominant trait may have parents that are both affected or only one is affected.
Heterozygous parents that expresses the dominant trait can have an unaffected offspring.
Autosomal Recessive Inheritance:
The trait have equal chance to appear on both female and male.
The trait CAN and USUALLY skip a row or rows (each row = a different generation) on the pedigree tree diagram.
If both parents expresses the recessive trait, all of the offspring must express the recessive trait.
Affected offspring expressing the recessive trait could (not necessary) have two unaffected heterozygous parents
Sex-Linked (X-Linked) Dominant Inheritance:
The trait have equal chance to appear on both female and male.
The trait CANNOT skip a row on the pedigree tree diagram.
Affected father will pass on the X-linked trait to the daughter(s) but NOT to son(s).
Affected mother will pass on the X-linked trait to both her daughter(s) AND son(s).
If the father is affected, all of his daughters will be affected. Also the affected father’s mother will be affected.
Generally, the trait will be expressed by females more often than in males.
Sex-Linked (X-Linked) Recessive Inheritance:
Generally, the trait will be expressed by males more often than in females.*
The trait can skip a row on the pedigree tree diagram.
Affected father will pass on the X-linked trait to the daughter(s) but NOT to son(s).
Affected mother will pass on the X-linked trait to both her daughter(s) AND son(s). If the mother is affected, the sons will all be affected, regardless of father being affected or not. Also, the affected mother’s father will be affected too (i.e. show the X-linked recessive trait)
If the mother the carrier (unaffected) and the father is unaffected, then the male offspring will have a 50% chance to be affected and daughters will not be affected (due to X allele from father).
* If the mother is carrier and the father is affected, then is equal chance of females and male being affected.
Of course, if the trait is Y-linked then it is easy as only males will be affected. This is because females do not have Y sex chromosomes.
It is important to note that you CANNOT PROVE that a trait is sex-linked using a pedigree!
That being said, pedigrees CAN be used to eliminate a method of inheritance (using patterns) as being possibly responsible for the transfer of trait from parent to offspring.
Constructing a pedigree
When constructing a pedigree, we use many symbols. Therefore, we must draw a legend or a key to inform the HSC Biology marker what each symbol that you drew means!
A circle typically refers to a female by convention.
A square typically refers to a male by convention.
A diamond is typically used to reference an individual with an unknown gender such as an unborn baby where you want to determine the baby’s likelihood in inheriting the trait (or gene) of concern.
A shaded circle or square would suggest that the individual is affected by the trait of concern. The trait of concern is usually recessive. The HSC question that you are given will let you know.
A half-shaded circle or square would suggest that the individual is a carrier of the trait of concern.
The actual pedigree structure itself:
The pedigree consists of rows.
Each row represents a different generation. Looking from top to bottom, the first row is the oldest generation.
Hence, the siblings of the same generation are aligned across the same row.
Each row or generation is labelled in roman numerals.
The first and oldest generation is labelled ‘I’ in roman numerals.
Individuals of the every generation are assigned a number from 1 and increases by an incremental of one. So, the oldest sibling is given the number “1”.
The second oldest sibling is given the number “2” and so on.
You draw a single horizontal line connecting the symbols of the two individuals to show the crossing over.
You draw a single vertical line in between the horizontal crossing over line that is connected to show the birth of a new generation (new horizontal line below the crossing over line). Remember that each horizontal line means a new generation!
Draw vertical lines extending down a generation horizontal line to depict offsprings of the parent.
A spouse from another family can be shown in the pedigree tree using a horizontal line that connect his or her partner.
Example of a pedigree
As illustrated in the small box in the bottom left, please note the correct way that you are to draw the vertical line that is extending from the the 2nd horizontal line. It should NOT be in line with the vertical line that is extending from the 1st horizontal line.
Looking at the actual pedigree, as illustrated in the diagram above, there is a point that is worth your attention. Specifically, the point is in regards to the diamond symbol.
The question mark in the diamond is to show you do not know the genotype of the individual (the individual is an unborn offspring in this case). Usually this is when you are not given sufficient information to work out the genotype of the individual. (Usually the question may want you to determine the genotype of this individual).
However, if a definitive genotype can be arrived using Punnett squares, you can re-draw the pedigree to include the correct shading of the diamond for the unborn offspring! Obviously, to definitively work out the genotype of the offspring, the question would require to provide you sufficient information of other individuals’ genotype that allows you to work out the offspring’s genotype. If not, you should include the probabilities of the different genotype combinations the offspring can inherit and you leave the pedigree as it is, i.e. with the question mark drawn in place as it is not conclusive of what the genotype of the individual is.
Learning Objective #3 - Collect, record and present data to represent frequencies of characteristics in a population, in order, to identify trends, patterns, relationships and limitations in data, for example:
- Examining frequency data
- Analysing single nucleotide polymorphism (SNP)
Single Nucleotide Polymorphism (SNP)
The term polymorphism refers to the presence of two or more alleles for a gene that exist in at least 1% of the species population.
An example of a polymorphism that we have explored already is the three alleles that governs the blood type for an organism – A, B and O alleles. Since these alleles exist in at least 1% of the species’ population genome, alleles for blood type is an example of polymorphism.
Fun fact: There are over 14 million examples of polymorphism in the entire human populations’ genome. Out of the 14 million, 10 million of them are due to single nucleotide polymorphism (SNP).
Other less common types of polymorphism that are beyond the HSC Biology Syllabus include protein polymorphism and restriction fragment length polymorphisms and copy number polymorphism.
Single nucleotide polymorphism is the scenario where one nucleotide in the genome of an individual is different to the rest of the population’s nucleotide at the same locus of the chromosome (or same location in the DNA or gene). This is because, due to SNP, a new nucleotide randomly replaces or substitutes an existing nucleotide in the organism’s DNA sequence.
For this reason, single nucleotide polymorphism is sometimes referred to as single nucleotide substitution.
This means that SNP will cause an organism to have a different nucleotide compared to the rest of the population which has the ‘normal’ nucleotide (normal is defined by population majority).
In case that you have forgotten, recall that the term ‘genome’ refers to all the genetic materials an organism has as a whole.
For a change in DNA sequence to be considered a SNP, the same SNP must be experienced by organisms in at least 1% or more of the species’s population. If not, the change in DNA sequence is deemed a mutation (which we talked about earlier).
For such reason, SNP events are far more stable and persist longer in the population than mutation where a mutation may be an one-off event (less than 1% of the population has it).
When SNPs (single nucleotide polymorphisms) occurs inside a gene, this would mean that the DNA sequence is altered and, hence, new allele are created. That being said, approximately 99% of all SNPs events occur outside the coding region of the DNA responsible for specifying for a protein.
This means that these SNPs that occur outside the coding region of DNA will NOT have a direct effect on the organism’s ability to function normally as the majority of SNPs mainly occur in the non-coding areas of the DNA. This is because DNA sequences located in the non-coding DNA sequences don’t specify for a protein.
The DNA sequences belonging in the non-coding region of DNA do have an indirect effect on the organism’s ability to function normally which you will learn in Module 6 as some are associated for the likelihood of the organism developing a disease.
The event of SNPs are not rare in humans, in fact, they occur approximately in every 100 to 300 nucleotides (300 on average) in a human’s genome which consists of approximately 3 billion nucleotides. Due to this, SNPs are responsible for over 90% of the genetic variation in the human population.
Even though SNP responsible for over 90% of the genetic variation in the human population, the actual total genetic variation between humans are 0.01% and about 90% of it are due to SNPs. The rest are due to mutation and other types of polymorphisms.
It is important to note that SNPs is NOT directly give rise to diseases, they could however increase or decrease the likelihood of the offspring developing a particular disease for the allele due to SNP that he or she inherited from her parent. Recall that if SNPs occur in the coding region of the DNA, it can create new alleles. These new alleles can be inherited by offsprings. One allele from each parent. Some of new alleles due to SNP events may be associated to increasing the likelihood developing a particular disease when inherited. To enhance you understanding, let’s look at an general example:
Suppose you have a gene responsible for a disease. Due to SNP, you have 3 different possible alleles in the population for such gene, let those be A, B and C. Suppose that if you inherit C from one of your parent, you will be more likely to develop a disease than if you inherited a B allele instead. That being said, the likelihood of developing the disease or trait is NOT absolute (i.e. not 100%) if you inherit allele C. An offspring can inherit two C alleles and not develop the disease.
That being said, the fact that SNPs influences the likelihood of an individual developing a disease or a trait is why SNP is a hot interest area of study for biologists currently.
There are generally two different groups of SNPs.
One group of SNPs (coding SNPs) is classified based on the SNPs which occur within the region of DNA (gene) that is responsible for protein coding.
The other group (non-coding SNPs) is classified based on SNPs that occur outside the region of DNA that is responsible for protein coding, i.e. SNPs that occur in genes (region of DNA or DNA sequence) that do not code for protein.
There are two types of SNPs that belong to the coding SNPs group.
These are synonymous SNPs and non-synonymous SNPs.
Synonymous SNPs – Substitution of nucleotide occurs. These changes the codon sequence but does NOT lead to a change in the amino acid sequence as the new codon sequence specifies the same amino acid. This is known as codon degeneracy where multiple codon sequence can code for the same protein. Synonymous SNPs therefore DOES NOT alter the amino acid sequence. The resulting protein structure and function will NOT be affected. This is because the altered gene sequence due to SNPs changed the mRNA codon. However, the mRNA codon still codes for the same protein.
Non-synonymous SNPs – Substitution of nucleotide occurs. These will also change the codon sequence but WILL lead to a change in the amino acid sequence as the new codon sequence specifies a different amino acid that is different to the ‘normal’ (or population majority). Non-synonymous SNPs therefore alters the amino acid sequence. This will affect the structure and function of the protein because SNPs the altered gene sequence (leading to a change in mRNA codon). The resulting protein may be non-functional.
Non-synonymous SNPs can be divided further into Mis-sense SNP and Nonsense SNP.
Mis-sense SNP – Substitution of nucleotide occurs. This will lead to an altered codon sequence which specifies a different amino acid.
Nonsense SNP – Substitution of nucleotide occurs. This will lead to an altered codon sequence which specifies for a stop codon. The more upstream direction which the SNP occurs on the DNA sequence, the earlier the stop codon will be specified during protein synthesis. This would mean that resulting protein created will be shorter (depending on how early or upstream which the Nonsense SNP substitution occurs in the DNA). The earlier which the nonsense SNP takes place, the more shorter the protein will be and the greater chance that the resulting protein will be non-functional.
SNPs are not always a disadvantage! Sometimes, SNPs can have rise to proteins with enhanced function, allowing the organism to better survive in its environment by tolerating against selective pressure. So maybe the organism will be real life spiderman? :O
Importance of SNPs
Although most SNPs occur outside the region of DNA that is responsible for coding proteins, we have already mentioned that SNPs within the regions of DNA that are responsible for coding protein could indeed lead to new protein structures and functions.
This means that SNPs can have an effect on altering the physical, physiological and even behavioural traits (e.g. personality) or characteristics such as of an organism.
Genome-wide association studies (GWAS) employs computer technologies to obtains a detailed report on the genome of individuals. Researchers scan the sections of DNA that have the greatest variance amongst species the population and documenting the SNPs of the scanned individuals.
By obtaining genome reports of individuals with and without a trait (or with and without disease) in the population, scientists can attempt to determine the genes that are responsible for certain traits or diseases. This is done by comparing the frequency of SNPs that appear between an affected individual and an unaffected individual. The general rule of thumb is that the higher the frequency of SNPs, the more likely those SNPs are related to the trait or disease the affected individual has developed.
As technological advances, powerful computers with more advanced algorithms at lower costs can be developed to assist scientists in unlock the secrets of the genes or allele combinations that determines certain traits or diseases! Also, with future technological advancements, a greater volume of data can be processed at a shorter period of time.
Limitations of SNPs
Recall in previous week’s notes, the organism’s exposure to ambient environment condition also plays a role in determining the phenotype of an organism not just genotype!
Genotype + Environment = Phenotype
Therefore, although SNPs can analysed by computer algorithm to determine whether SNPs (and thus the affected gene) is responsible for an individual developing the concerned trait (or disease), scientists would still require to take into account of the environment factors that may contribute to an individual developing the trait (or disease) of concern.
Some environmental factors may alter gene expression such as the individual’s lifestyle and diet which may make the organism more prone to developing a trait or disease.
Diseases can also be monogenic or multigenic. Monogenic are caused by one ‘faulty’ gene whereas multigenic is caused by multiple ‘faulty’ gene. Using SNPs to identify multigenic diseases are much harder as multiple genes are responsible for the individual to develop such disease or trait.
That being said, advancements in technologies could allow scientists to definitively associate all of the SNPs that are responsible for the development of a disease or trait. This will assist in the development of medicine to prevent the development of diseases or unfavourable traits because scientists would have a greater understanding how the affected individual will react to medicine treatments and cure/prevent the disease.
Examining frequency data
Genetic variability contributes to genetic diversity.
Genetic diversity in a population refers to the sum of all traits in a population.
The greater the genetic diversity a population has, the more capable it is able to adapt to changing selective pressures in the environment which the species residues in.
Population genetics involves the study of the frequency of alleles for one or more genes in a population due to changes in selective pressures in the environment over time. The dependent variable of the study would be the frequency of alleles for a gene. The independent variable would therefore be a stimulus or a change in environmental condition such as a temperature change.
Mathematical models are often used to predict the change in allele frequency of a gene due to a change in one environmental condition such as temperature. These model allow scientists to obtain results which they can use to compare against actual or real data such as those obtained from observational studies. These real data or empirical data will allow scientists to confirm the conclusions about allele frequency due to a change in a particular environmental factor which they have drawn from using their mathematical models.
Genetic variability in terms of allele frequency can be determined using mathematical expressions.
For example, suppose you wish to determine the frequency of an allele that codes for blue eye colour in unicorns. You would have to obtain the total number of alleles that codes for blue eye colour in the unicorn population and divide it over the total number of all alleles that codes for eye colour in the unicorn population.
Frequency of blue – eye colour allele = Number of blue-eye alleles in unicorn population / Total number of colour-eye alleles in unicorn population
The above expression is oversimplified. This is because we are using the phenotype of organisms in the population to determine allele frequency. If you recall, there are dominant and recessive alleles. Suppose that if blue eye colour is specified by a dominant blue eye colour allele (A) and the complementary recessive green eye colour allele is (a).
In such scenario, the blue-eye colour unicorns may have genotype of AA or Aa. This cannot be determined by simple sampling techniques such as simple observation. We need to use:
Frequency of A (dominant blue eye colour allele) Allele = (2 x number of AA alleles + number of Aa alleles) / 2 x number of alleles coding for eye colour in the sample.
Gregor Mendel
Gregor Mendel studied pea plants and performed a lot experiments with them via artificial pollination.
He established pure-breed parental generation line. Pure breed species are species that Mendel knows that their alleles are homozygous for a particular trait. For example, a pure-bred yellow pea plant will have two alleles (homozygous) that specifies its yellow seed colour.
Mendel established pure-breeding lines for pea plants with round (RR) and wrinkled (ww) seeds which he used as his parental generation as shown on the left in the diagram below.
He also established pure-breeding lines for many other traits in pea plants such as stem height, etc.
What he found was the allele that coded for round pea plants were dominant over the alleles that specified for wrinkled pea plants. This is because he observed that 100% of the offspring in the first generation (F1) were had round peas.
After crossing over heterozygous dominant offsprings in the first generation (RW x RW), he found that 75% of the pea plants were round and 25% of the pea plants were wrinkled in the second generation (F2). This was a 3:1 ratio.
This indicated that wrinkled pea plants was a recessive allele and will only be expressed if the organism’s genotype consists of the two recessive alleles coding for a particular gene. This is because no dominant allele will be present to ‘override’ the recessive allele.
These observations from the first and second generation pea plants became evidence to support Mendel’s proposal of the law of dominance in alleles for specifying a gene.
Mendel also established pure-breeding lines for yellow (YY) and green (gg) coloured pea plants which he used as his parental generation as shown on the right in the diagram below.
Essentially, that approximately 75% of the second generation (F2) pea plants had the dominant yellow trait.
In his experiments, Mendel also proposed the law of independent assortment as well as the law of segregation which we have talked about in meiosis.
However, we will revisit the law of independent assortment again as we look into dihybrid crosses.
Extension Content on Inheritance
(Dihybrid Crosses)
When we explored monohybrid crosses in our Punnett Square examples earlier, that is, we only dealt with parents that have different alleles for the same, single trait.
Now, we will explore dihybrid crosses which means that it will parents that have different alleles for two traits. This means that in our Punnett Square, we need to consider alleles for two traits at the same time.
This is not hard to do. Keep in mind that the offspring’s inheritance of alleles for a gene is independent to the offspring inheriting an allele for a different gene.
For example, suppose that the offspring can either inherit a blue or black allele for eye colour and a brown or black allele for hair colour. If the offspring inherited two blue alleles for his or her eye colour, one from each parent, such genotype (OR any other genotype for eye colour – e.g. one blue and one black eye colour allele) will NOT affect the offspring’s chances of inheriting brown or black allele for its hair colour gene.
Recall that from Mendel’s pea plant experiment, the observation he made in his second generation pea plants were that 3/4 of his pea plants were yellow and 1/4 of his pea plants were green. Similarly, 3/4 of his pea plants were round and 1/4 of them were wrinkled. These two sets of result, one for each gene, was determined using monohybrid cross was shown previously in this week’s notes.
Suppose that in the exam, you have a question that asks what is the probably of a plant in the F2 (second generation) that is both yellow and round? Well, you can use a draw a Punnett Square to show the dihybrid cross of the F1 parents to form the pea plants in the F2 generation.
OR, you can simply use the product rule for independent events in mathematics to calculate the probability that a F2 pea plant will be both yellow and round. That is,
Probably that the F2 plant will be yellow and round = Probability (F2 plant is yellow) x Probability (F2 plant is round) = 3/4 x 3/4 = 9/16.
We only know the probability of F2 plant is yellow (3/4) and the probability of F2 plant is round (3/4) because we did one monohybrid for the pea colour for two F1 generation and one monohybrid cross for the shape of the pea plant for two F1 generation plants.
What if we don’t want to construct two monohybrid crosses, one for each trait?
Well, another method is that we can construct a dihybrid cross and you still get 9/16 as an answer as shown below.
Notice from the dihybrid cross over, the amount of F2 pea plants that are round and yellow is 9/16, as shown by the red check marks.
The reason why the result of doing two Punnett Square monohybrid crosses is the same as the result of one dihybrid cross (both is 9/16) is due to Mendel’s law of independent assortment.
Recall that independent assortment occurs during Metaphase I. The effect independent assortment is that each of the two haploid cell may inherit different alleles.
This means that if scenario #1 occurs, one haploid cell will inherit tt alleles for height after Telophase I and the other haploid cell will TT allele for height after Telophase I. Each of these cells then undergo meiosis II and further divides into two gametes.
If scenario #2 occurs, the one haploid cell would inherit TT alleles for height instead of tt (in scenario one) after Telophase I. This would mean that other haploid cell would inherit tt allele for height instead of TT in scenario #2.
The fact that alleles specifying for different genes of non-homologous chromosome assort themselves independently during metaphase I means that there is an increase the genetic variation of the alleles which the gametes can inherit at the end of meiosis II.
Because of independent assortment, you can use the product rule as shown earlier. This is why the results of using one dihybrid Punnett Square yields the same results as using two monohybrid crosses then multiplying the results of each monohybrid crosses together. That is, the probability of inheriting alleles for one trait (or gene) does not affect the probability of the same offspring inheriting alleles for a different trait.
Week 4 Homework Questions
Homework Question #1: Explain the significance of crossing over in contributing to the offspring’s genotype
Homework Question #2: Explain the significance of fertilisation in contributing to the offspring’s genotype
Homework Question #3: Explain the significance of mutation in contributing to the offspring’s genotype
Homework Question #4: Explain the difference between autosomal, sex-linked, co-dominance, incomplete and multiple-allele inheritance of alleles by the offspring. Provide an example of each. (HINT: Explain in terms of the nature of alleles and/or where their location on chromosomes in the case of sex-linkage)
Homework Question #5: Explain whether or not each of the modes of inheritance in the previous question obeys mendelian inheritance.
Homework Question #6: Describe the term ‘single nucleotide polymorphism’
Homework Question #7: Explain three limitations of single nucleotide polymorphism
Homework Question #8: Describe the advantages and limitations of using pedigrees
Week 4 Curveball Questions
Curveball Question #1: Describe the difference between mutation and single nucleotide polymorphism
Curveball Question #2: The father with black eyes and mother with green eyes have two children with black eyes and one children with green eye. Draw all the possible Punnett Square to account for the possible genotypes of the parents and their children. In your answer, state the genotypes of the father and the three children. Assume in this question that black eye colour (B) is dominant over green eye colour (b)
Curveball Question #3: A homozygous tall exotic plant crosses over a homozygous short exotic plant. Assume that tall exotic plants (T) are dominant over short exotic plants (t). Construct a Punnett Square to illustrate the possible phenotype and genotype of the offspring when the two exotic plants are crossed over.
Curveball Question #4: Suppose a recessive trait will be expressed in the individual if the individual has aa genotype. Suppose that four percent of the population have the recessive ‘a’ allele and the reminder of the population have the dominant ‘A’ allele. Construct a Punnett Square to determine the different genotype that the offspring will have when two heterozygous dominant parents (Aa) are crossed over.
Curveball Question #5: Suppose that unicorn A is crossed over with three different unicorns (unicorn B, C and D). The offsprings when unicorn A is crossed over with B, C, D produced unicorns that have rainbow, white and white hair respectively.
Suppose that unicorn A has rainbow hair. Unicorn B, C and D have white, white and rainbow hair respectively.
What are the genotypes of unicorn A, B, C and D?
Curveball Question #6: Suppose that a unicorn’s rainbow hair colour is specified by R allele in the unicorn’s genotype for hair colour. This means that r allele specifies purely white hair colour in the unicorn population. The genotype RR is so powerful that if an unicorn zygote has such genotype, it will not successfully develop into a foetus. Due to this, there will not be a single unicorn with RR genotype in the existing population. Suppose that Kenneth was having fun one day and he crossed over two rainbow unicorns each with rainbow hair. Kenneth then counted the number of new unicorn that was born due to the crossing over.
Hair colour | Number of offspring
Rainbow = 40
White = 20
(a) Construct a Punnett Square to illustrate the crossing over of the two rainbow unicorns which Kenneth have chosen
(b) From the results in part (a), what the percentage of unicorns has at least one R allele in its genotype
(c) Account for the reason the percentage of rainbow unicorns from the Punnett Square result is different from the number of rainbow unicorns that was observed by Kenneth.
(d) Suppose that Kenneth found a magical potion and gave it to all the unicorns so that offsprings with the NN genotype can exist in the unicorn population. After this, Kenneth crossed over two heterozygous unicorn parents and counted the resulting offsprings. He counted 4 unicorns and recorded their phenotype. All four of them were rainbow. Construct a Punnett Square for this crossing over and propose a reason to why the results of your Punnett Square is different to what Kenneth observed in the unicorn population.
Research Exercise: Research one disease that is associated to SNP!